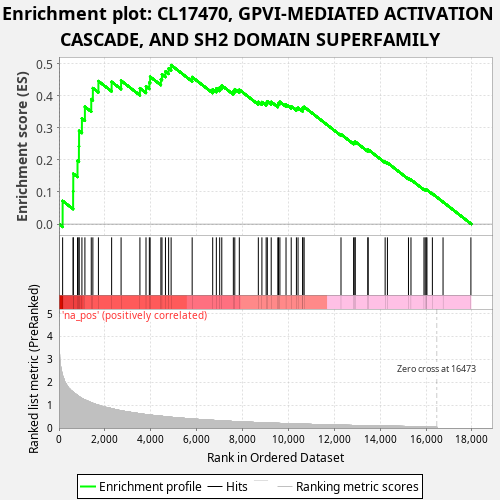
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Irritable bowel syndrome__20002-both_sexes-1154 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL17470, GPVI-MEDIATED ACTIVATION CASCADE, AND SH2 DOMAIN SUPERFAMILY |
| Enrichment Score (ES) | 0.49547958 |
| Normalized Enrichment Score (NES) | 1.0933608 |
| Nominal p-value | 0.23 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | PIK3R3 | 158 | 2.283 | 0.0722 | Yes |
| 2 | BTK | 616 | 1.562 | 0.1021 | Yes |
| 3 | FYN | 622 | 1.556 | 0.1571 | Yes |
| 4 | PIK3CB | 807 | 1.422 | 0.1973 | Yes |
| 5 | SHC3 | 868 | 1.371 | 0.2426 | Yes |
| 6 | PTPN1 | 875 | 1.363 | 0.2907 | Yes |
| 7 | IRS2 | 999 | 1.286 | 0.3295 | Yes |
| 8 | VAV3 | 1129 | 1.223 | 0.3657 | Yes |
| 9 | FGR | 1406 | 1.102 | 0.3894 | Yes |
| 10 | LAT | 1476 | 1.075 | 0.4237 | Yes |
| 11 | PTPN11 | 1718 | 0.995 | 0.4456 | Yes |
| 12 | ZAP70 | 2294 | 0.844 | 0.4434 | Yes |
| 13 | UBASH3A | 2706 | 0.749 | 0.4471 | Yes |
| 14 | PIK3R1 | 3527 | 0.621 | 0.4233 | Yes |
| 15 | PLCG2 | 3792 | 0.582 | 0.4293 | Yes |
| 16 | TXK | 3938 | 0.564 | 0.4412 | Yes |
| 17 | SOCS4 | 3972 | 0.560 | 0.4593 | Yes |
| 18 | PIK3CA | 4441 | 0.511 | 0.4513 | Yes |
| 19 | IGF1R | 4488 | 0.507 | 0.4667 | Yes |
| 20 | ITK | 4640 | 0.491 | 0.4757 | Yes |
| 21 | BCAR1 | 4778 | 0.477 | 0.4850 | Yes |
| 22 | PIK3R5 | 4887 | 0.464 | 0.4955 | Yes |
| 23 | GRAP2 | 5804 | 0.392 | 0.4582 | No |
| 24 | GAB1 | 6699 | 0.332 | 0.4201 | No |
| 25 | CD79A | 6858 | 0.323 | 0.4228 | No |
| 26 | SHC1 | 7003 | 0.316 | 0.4259 | No |
| 27 | VAV2 | 7095 | 0.311 | 0.4319 | No |
| 28 | LCP2 | 7599 | 0.281 | 0.4138 | No |
| 29 | DAPP1 | 7662 | 0.278 | 0.4202 | No |
| 30 | INSRR | 7859 | 0.269 | 0.4188 | No |
| 31 | VAV1 | 8688 | 0.232 | 0.3808 | No |
| 32 | CBL | 8844 | 0.226 | 0.3802 | No |
| 33 | SLA2 | 9029 | 0.219 | 0.3777 | No |
| 34 | BCR | 9080 | 0.217 | 0.3826 | No |
| 35 | PIK3AP1 | 9249 | 0.211 | 0.3808 | No |
| 36 | SYK | 9539 | 0.200 | 0.3717 | No |
| 37 | PIK3CD | 9568 | 0.200 | 0.3773 | No |
| 38 | CRKL | 9628 | 0.198 | 0.3810 | No |
| 39 | PIK3R2 | 9895 | 0.189 | 0.3728 | No |
| 40 | SHC4 | 10121 | 0.181 | 0.3667 | No |
| 41 | GAB2 | 10348 | 0.173 | 0.3603 | No |
| 42 | CRK | 10419 | 0.171 | 0.3624 | No |
| 43 | PLCG1 | 10623 | 0.165 | 0.3569 | No |
| 44 | IRS1 | 10625 | 0.164 | 0.3627 | No |
| 45 | PTK2 | 10684 | 0.162 | 0.3653 | No |
| 46 | RAPGEF1 | 12291 | 0.120 | 0.2799 | No |
| 47 | INSR | 12840 | 0.108 | 0.2531 | No |
| 48 | FRS2 | 12897 | 0.107 | 0.2538 | No |
| 49 | LYN | 12908 | 0.107 | 0.2570 | No |
| 50 | GP6 | 13461 | 0.096 | 0.2296 | No |
| 51 | IRS4 | 13473 | 0.095 | 0.2323 | No |
| 52 | SOS2 | 14215 | 0.080 | 0.1938 | No |
| 53 | GRB2 | 14315 | 0.079 | 0.1911 | No |
| 54 | TEC | 15232 | 0.064 | 0.1422 | No |
| 55 | INS | 15341 | 0.061 | 0.1384 | No |
| 56 | PDGFRB | 15908 | 0.050 | 0.1085 | No |
| 57 | SRC | 15960 | 0.048 | 0.1074 | No |
| 58 | LAT2 | 16005 | 0.047 | 0.1066 | No |
| 59 | BLNK | 16037 | 0.046 | 0.1065 | No |
| 60 | CD79B | 16274 | 0.040 | 0.0948 | No |
| 61 | PIK3CG | 16739 | -0.000 | 0.0689 | No |
| 62 | PIK3R6 | 17953 | -0.000 | 0.0012 | No |